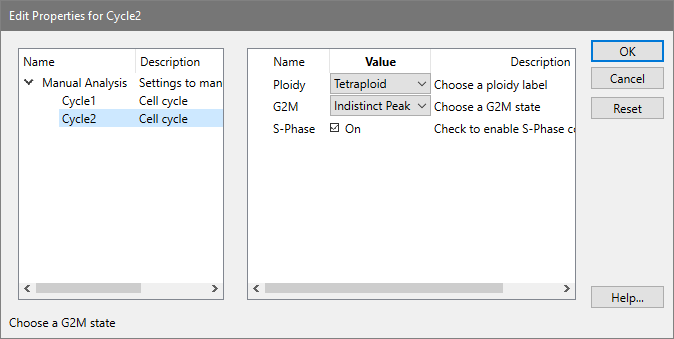
These options appear when you chose a Cycle in the tree on the left side of the Edit Properties for Manual Analysis dialog box. The options allow you to fine-tune the properties for a set of components used to model one cell cycle in the model.

Ploidy
This option allows you to select a different ploidy label for the cell cycle. The model template you select in the Edit Properties for Manual analysis dialog determines the default value for this option.
Range labels and results reported on the report are based on the ploidy names assigned to cell cycles.
G2M
There are three options available for the way G2M is modeled: Disabled, Visible peak, and Indistinct peak. When Disabled is selected, no G2M will be modeled for the cycle. Choosing Visible peak allows the program to adjust the mean position of the G2M component for the best fit, the lowest reduced chi-square (RCS). If the peak for G2M is not clearly defined, choose Indistinct peak. With this setting, the position of the G2M is based on the G1 position multiplied by the linearity factor.
S-Phase
This checkbox allows you to enable or disable the S-Phase component for the cycle. Normally, you should disable an S-Phase if another more important cycle is significantly overlapping this cycle. For example, in the near-Diploid model, the Diploid S-Phase is disabled because the DNA Index (DI) for the Aneuploid cycle is less than 1.3 and therefore the S-Phases are virtually superimposed on one another.