

Overview
The Quick Model Editor allows you to make changes to the current model. It is available when the model you are using is created by AutoAnalysis or by using the Choose Model option.
Changes you make with this dialog are applied to the model when you click OK. In order to see the changes to the model, you need to re-analyze the data by clicking the Fit button or choosing Execute Model from the Analysis menu.
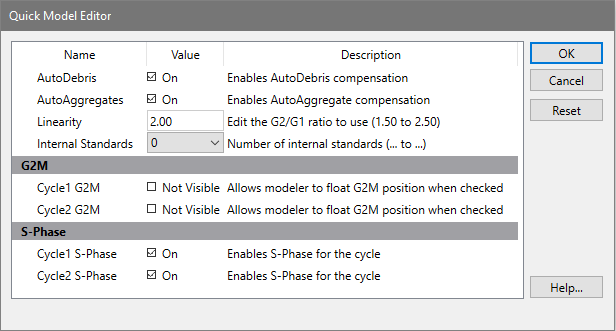
AutoDebris
This checkbox determines whether the AutoDebris model component is active or not. When checked, AutoDebris is activated. The AutoDebris component is used to model debris in data. It can be left active even when samples display little or no debris, because its shape and amplitude are based on probability distributions unique to each sample.
AutoAggregates
When this option is checked, the AutoAggregates model component is activated. This component is used to model the continuum of aggregate forms found in many samples. It can be left active even when there is little or no aggregation in the sample, because its shape and amplitude are based on probability distributions unique to each sample
Linearity
This field modifies the appropriate G2M dependency settings and modifies the aggregate model component's linearity setting. When the user indicates that a G2M is not visible, the program makes the position and standard deviation of the G2M model component dependent on the G1 by this linearity setting. It updates the linearity setting for the aggregate model component as well.
Internal standards
This option determines the number of peaks attributed to standards. Standard peaks typically occur below the first G1 peak. However, you can configure the expected locations for standards in your samples using the Auto Analysis Settings dialog box.
G2M Section
The options in this section of the dialog allow you to control how the G2M for each cycle is modeled. If the option is checked (Visible), the program will use the associated G2M range to estimate the position and it will allow that position to be adjusted by the modeling process. If the option is not checked, the position of the G2M will be computed by multiplying the G1 position by the linearity value.
Generally, if a G2M is a well-defined peak that does not overlap another peak, you can enable the option and the model will provide a more accurate result. However, if the G2M peak is not clearly defined or if it substantially overlaps another peak, uncheck the option. If the option is checked when a peak is not well defined, the model will produce less accurate results.
S-Phase Section
The options in this section control the state of the S-Phase model component for each cell cycle in the model. If a check appears in the checkbox, the S-Phase component is active and the program will attempt to model it. Clear the check to disable the S-Phase component for the model.
In samples such as near Diploid histograms, the S-Phases of two or more cycles can be significantly overlapped. This can create problems for the modeling process. In these cases, it is better to disable one of the S-Phase components, normally the Diploid S-Phase.
OK
Apply the settings to the current model and put away the dialog box.
Cancel
Close the dialog and discard any changes you have made.
Reset
This option resets the the properties in this dialog to their default states.