

Use this command to manually adjust the ranges for the model.
ModFit LT uses specific ranges to identify important parts of a histogram, usually peaks. Automatic Analysis positions ranges for you. However, if you need to correct a range position or if you manually select a model for analysis, you will need to position ranges yourself.
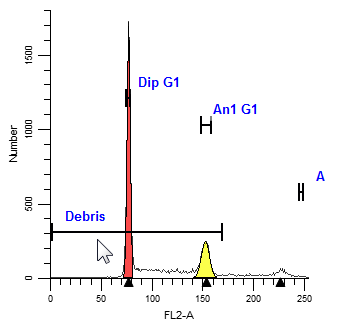
The rule of thumb for positioning ranges is to make sure each range identifies one, unique peak. Ranges should be positioned so that they do not overlap one another. When a range is positioned correctly, you will see the associated portion of the histogram filled in with a color indicating the range estimate.
The exception to the rule for placing ranges is shown in near-Diploid histogram. In these histograms, you may need to place ranges so that they are not centered on the peaks. Instead, move the ranges slightly apart to help differentiate the two, overlapping peaks.
Only move or resize ranges that need to be adjusted. By leaving correctly positioned ranges alone, you help improve user-to-user consistency.
The vertical position of ranges has no impact on the range estimate. You can arrange ranges vertically so that they are easy to read and access.
Ranges and their uses
Debris |
This range provides an estimate of debris. The left side should be placed at the highest portion of the beginning of debris. The right side should extend approximately two-thirds of the histogram scale. The vertical position has no impact on the estimate. |
Stnd1 |
This range is used to identify the first internal standard, if there is one. If you have named the internal standard, the name of the range is changed to the name of the standard. Center the range over the first standard peak. |
Stnd2 |
This range is used to identify the second internal standard, if there is one. If you have named the internal standard, the name of the range is changed to the name of the standard. Center the range over the second standard peak. |
Apop |
This range identifies the apoptosis peak, if you have enabled the apoptosis component. The range should be centered over the peak. |
Dip G1 |
This range identifies Diploid G0-G1 peak. The range should be centered over the G1peak. |
Dip G2 |
This range identifies Diploid G2-M peak. The range should be centered over the G2 peak. |
An1 G1 |
This range identifies an Aneuploid G0-G1 peak. The first Aneuploid cycle will use An1, the second An2, and so forth. The range should be centered over the G1 peak. |
An1 G2 |
This range identifies an Aneuploid G2-M peak. The first Aneuploid cycle will use An1, the second An2, and so forth. The range should be centered over the G2 peak. |
G1 |
This range is used by the Sync Wizard to identify the Diploid G0-G1 peak. The range should be centered over the peak. |
Parent |
This range is used by the Cell Tracking Wizard to identify the Parent population. |
Gen1 |
This range is used by the Cell Tracking Wizard to identify a daughter generation population, if the Floating option is selected on the Generations tab of the wizard dialog. Gen1 is the generation closest to the parent, Gen2 is to the left of Gen1, etc. |